Eli Broemer
/E·Lye Bray·mer/
Abstract
Full PDFDevelopment of a Method to Quantify Elastin Content in Large Datasets of Bladder Tissue Histology with Machine Learning Algorithm
Eli Broemer (1), Sara Roccabianca (1)
(1) Mechanical Engineering
Michigan State University
East Lansing, MI, USA
Introduction
Extracellular matrix remodeling in which the structure and/or distribution of collagen and elastin fibers in the urinary bladder wall are modified has been associated with dysfunction [1]. Histological images are useful in studying microstructural changes, however, quantifying elastin content in these images is challenging. Specifically, the Verhoeff-van Gieson's (VVG) stain is known to effectively differentiate elastin fibers with sharp intense contrast, although it also stains cell nuclei and other connective tissue [2]. Despite having limitations, the stain is relatively easy, and elastin has high affinity to the dye, making VVG one of the most common elastic tissue stains [3].
Analyzing amounts of elastin in bladder tissue with VVG histologies has long been done qualitatively [4, 5]. Yet, quantitative attempts with VVG micrographs are less common, and may include visually scoring fiber density [6, 7], or estimating area fraction in a region of interest by thresholding the dark fibers from the light background [8, 9]. The primary limitation with the simple thresholding scheme is that cell nuclei and other noise are not removed by the cutoff value, or more strict cutoffs significantly erode the elastin fibers. To solve this issue, prior studies relied on substantial manual postprocessing and homogenization of connective tissue [8, 10]. Alternatively, computationally intense methods have been proposed, however limitations of these are that they either use proprietary software [10] or unsupervised image transforms to segment fibers [11]. Our goal is to improve on the latter approach by simplifying the segmentation procedure and by adding a machine learning postprocessing step to reduce the manual processing of large data sets.
Random forest [12] is a type of machine learning algorithm, and is regarded one of the best overall for prediction problems [13]. In short, the model fits an output dataset by building a decision tree used to interpret the input parameters [14]. Then, bootstrapping the data produces a forest of decision trees, and its final prediction is typically the average or most frequent result from the forest. This makes the algorithm robust to outliers, missing data, and irrelevant parameters. Random forest is easy to implement, fast to train, and has been applied to histology problems before [15]. Yet, its application to identify elastin in bladder tissue is novel and presents unique challenges.
The current work was initiated with a proof-of-concept by training a random forest algorithm on a pseudo dataset representative of VVG stained bladder tissue after simple thresholding to ensure feasibility. Then, we developed a software for processing true histology specimens. The software is used to, first, segment histology images and, second, to label the segments as elastin fibers or not. A segmenting protocol as simple as thresholding was enhanced by preprocessing more powerful image transforms. Then, the labelling process allows for multiple researchers to easily train a new elastin classification algorithm. When the random forest is sufficiently trained by showing reliable accuracy, it can automatize the quantification of elastin area fractions in large data sets, which will significantly improve the time needed for analysis.
Methods
A set of 35 VVG stained rat bladder wall microscopy images was contributed from a separate study for use. Image preprocessing starts when images are uploaded to the custom-made software. Each image is treated with a combination of transformations that were found to be generally successful at isolating the specific components of interest and masking the background noise. For every image, a set of 50 masks with varying levels of strictness is obtained by tweaking the combination of image transforms. Then, segmentation is done by a researcher choosing the strictness of the final mask with a slider to best capture elastin and remove noise, as shown in Figure 1. Combining varying image transforms (i.e., thresholding, contrast normalization, dilation, noise reduction) and then sorting the resulting image masks by the number of pixels masked allows the researcher to leverage these powerful techniques. This protocol was implemented into a web application using the python Bottle library. The software is hosted locally in the research lab, and lab members can login to easily segment images with this enhanced thresholding method, or label objects extracted from images.
Once an image is segmented, each object isolated by the mask is extracted, and on a different screen, shown in Figure 2, a researcher can label the objects based on their expertise. Furthermore, objects may carry labels from multiple researchers, and this is in fact encouraged as some objects are ambiguous and multiple opinions will reduce bias.
Results
The segmentation screen (Figure 1) allows quick masking of cells and elastin by utilizing preprocessed image transformations that are more powerful than simple thresholding. The mentally taxing process of tweaking the image transformations is supplanted with simply selecting more/less masking from the set of preprocessed transforms. Segmentation is faster and requires less image analysis intuition.
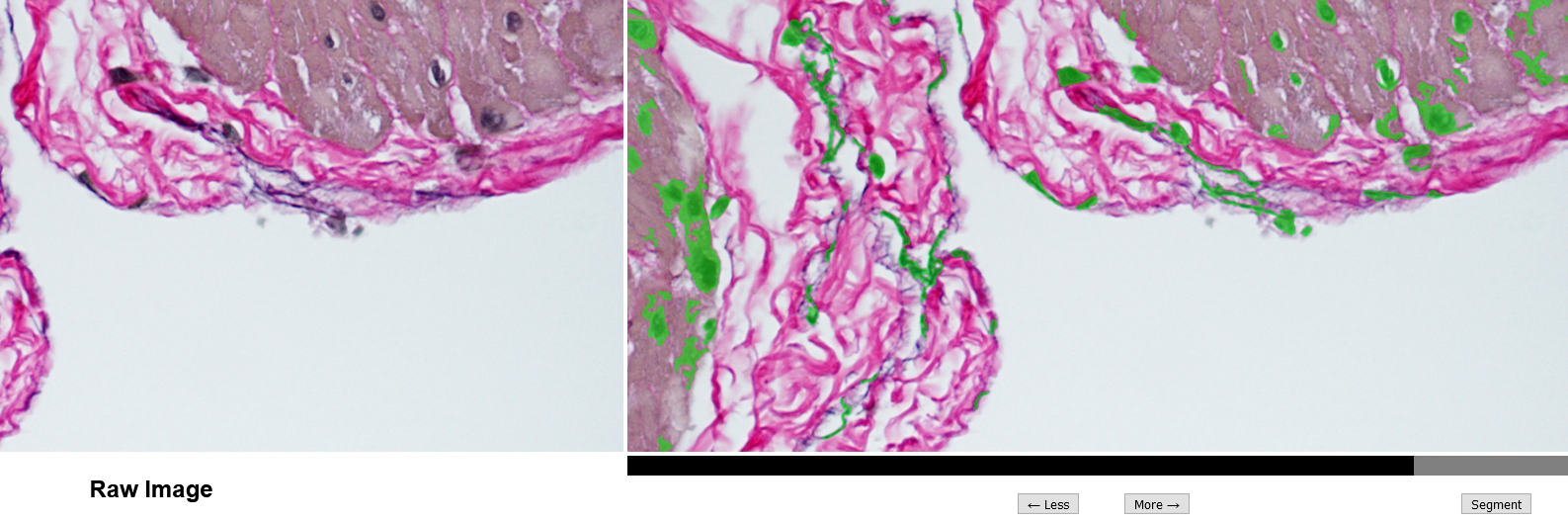
Figure 1: Segmentation screen utilizing enhanced thresholding method. Black/gray slider at the bottom has been adjusted so that cells and elastin are masked (green highlights) as best possible.
The labelling screen (Figure 2) presents individual microstructure for the researcher to identify. A given histology image may have between 50 - 200 objects isolated after segmentation, and it can take a time near 5 - 10 minutes to fully label an image. Since the software is hosted locally, loading and saving times are often near instant. This approach is faster and easier than the proof-of-concept method in which image files of isolated objects were labelled correspondingly in a spreadsheet. Data labels are used to train random forest predictions.
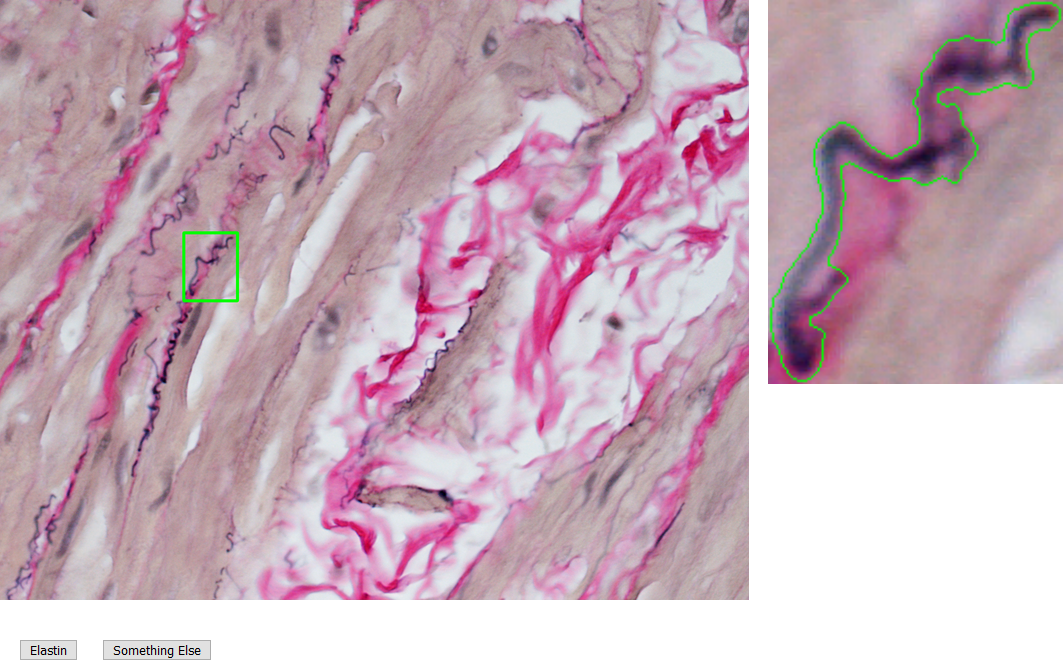
Figure 2: Labelling screen prompts user to identify if an object is elastin. Extracted object is shown with context (left) and also magnified with outline (right) for clarity.
Discussion
This histological elastin extraction software shows promise for estimating area fraction of elastic fibers in VVG stained tissues. Other means of quantifying elastin in tissues include commercial assays or Western blots. However, these procedures consume the sample and so the tissue morphology cannot be assessed from the same sample. Scanning electron and confocal microscopy are also used to capture images of elastin, although these technologies are more cost prohibitive.
Current limitations of this study are microstructure components that are not fully separated during segmentation. For example, an elastin fiber intersecting a cell are both joined as one object and cannot be labelled accurately. Next steps include solving this limitation by developing a secondary segmentation option when a researcher encounters this scenario during labelling.
As the development dataset is small, more histology images from bladder tissue are to be added. Then, a new random forest algorithm will be trained and tested on the data using this software. Fully labelled data is to be split 80/20 for tuning algorithm hyperparameters. That is, 80% of the data is used to train and 20% is used to test the algorithm. Random forest parameters including number of trees and branch splitting criteria will be optimized with 5-fold cross validation to assess accuracy. The model will also be tested on new data to further validate.
Later steps include implementing and comparing the accuracy of other machine learning schemes such as Support Vector Machine and Naive Bayes classifiers.
Acknowledgements
We thank fellow lab member Tyler Tuttle for the histology images, and the MSU Investigative Histopathology Lab for staining tissues. This research was inspired by Miguel Contreras' work at SB3C 2020 [16].
References
[1] Wognum, S et al., J Biomech Eng, 131(10):101018, 2009. [2] Verhoeff, FH, JAMA, L(11):876-877, 1908. [3] Carson, F & Cappellana CH, Histotechnology 4th ed., 2014. [4] Ji, P et al., World J Urol, 13:191-94, 1995. [5] Lemack, GE et al., Neurourol Urodynam, 18:55-68, 1999. [6] Kullmann, FA et al., Front Syst Neurosci, 2018. [7] Cantiello, F et al., Urology, 81(5):1018-23, 2013. [8] Jinrok, O et al., J Orthop Res, 22:1310-15, 2004. [9] Johal, N et al., J Pediatr Urol, 2020. [10] Karam, JA et al., BJU Int, 100(2):346-350, 2007. [11] Paul, K et al., Appl Mater Today, 22:100890, 2021. [12] Breiman, L, Mach Learn, 45:5-32, 2001. [13] Caruana, R & Niculescu-Mizil, A, ICML 2006, 161-168, 2006. [14] Zhang, C & Ma, Y, Ensemble Machine Learning, 2012. [15] Ikeda, T et al., J Hepato-Bil Pan Sci, 2020. [16] Contreras, M, Bachman, W & Long, D, SB3C, Abstract 466, 2020.
SB3C2021
Summer Biomechanics, Bioengineering, and Biotransport Conference
June 14-18, Virtual
 Home
Home